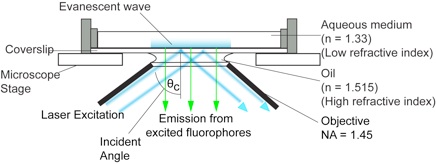
Most molecular imaging approaches right now use 'TIRFM' imaging, illustrated above. This involves illuminating a sample at the interface between 2 media - in this case oil (on the lens) and cells in medium. In this case, the laser light used to illuminate the sample is totally internally
Molecular imaging encompasses a variety of imaging techniques which between them, allow the visualisation, localisation (this page, and here), tracking and quantification of interactions between single molecules. We use all these techniques to quantify the dynamic behaviours of molecules inside living cells.
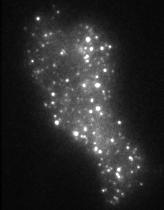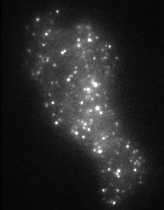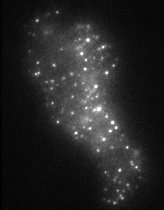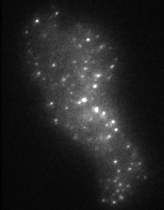
The movie on the right shows molecules of green fluorescent protein (GFP) inside single secretory vesicles at the membrane of a neuroendocrine cell. Each vesicle is 400 nm in diameter, and contains probably 100's of GFP molecules, making them clearly visible. Although this isn't 'molecular imaging' per se, it does demonstrate the techniques required to make the visualisation of single molecules possible. The advantages of TIRFM imaging are numerous; the cells can withstand the laser illumination for a long time, as most of the light doesn't penetrate into the sample. This allows us to image very quickly (30-40 frames per second, using an EM-CCD camera). The resulting image data are very high contrast, as the out-of-focus data are minimised by the thin illumination section.
Fast frame rates (on the millisecond scale, or faster) are required if we want to track objects moving inside cells. Clearly, if something is moving around, we need to image it faster than it moves in order to acquire accurate data. Even presenting the data has problems - how does one show the vast amount of information contained in these experiments, 'on paper'? The track on the left is from a single secretory vesicle (one of the spots from the movie, above). The track changes colour with time since the beginning of the experiment. We can colour-code the tracks to communicate speed, displacements, momentum etc, as well as generating vast arrays of numerical data. These approaches lay the foundations for experiments tracking the dynamic behaviours of single molecules inside cells.
LSI Molecular imaging - introduction
Images and data copyright LSI Laboratory
Introduction PALM Single particle tracking PALM FCS FLIM GSD Microscopy
As the behaviour of a single molecule in a cell is likely to be stochastic - that is, apparently random in isolation - it is desirable to examine large cohorts of single molecules all at once in a sample. The advantages of molecular imaging are clear - molecules are the smallest functional units in a cell, so determining how they behave can, in turn, tell us how cells function - and what goes wrong in disease.
reflected - the top surface of the glass coverslip where the cell is growing acts as a 'mirror', reflecting the light. In fact, a small amount of light does penetrate into the aqueous media - but only for a few 100 nanometres. This very thin section of illumination makes molecules at the very base of the cell visible.
