
Real time analysis of SNARE protein dynamics during exocytosis (MRC, to 2014)
The SNARE proteins are essential for secretion, and so normal physiology. Unfortunately, the techniques required to analyse the function of proteins at the molecular level in living samples have been severely limited until recently, so our understanding of their functions, locations and dynamic interactions remains limited. This project will address these questions, analysing SNARE protein molecular dynamics in living neurons. A combination of quantitative in vitro biochemistry and advanced imaging approaches will tell us how the proteins’ functions are regulated by calcium, in real time, with high spatial accuracy. Researchers: Rory Duncan, Colin Rickman, Deirdre Kavanagh, Ali Dun.
Molecular spatio-temporal dynamics of neuronal Sec-1 (Wellcome Trust, to 2014)
nSec-1, also known as Munc18-1, is one of only 4 proteins absolutely essential for secretion in cells. As secretion is central to normal physiology (for example, it is faulty in diabetes, schizophrenia, epilepsy etc), understanding how these proteins work is a key aim. Biochemists have told us about the proteins involved in secretion and their interactions, but the ‘wheres’ and ‘whens’ of the actions of these molecular machines in cells remain largely speculative. This project will determine where munc18 acts in a living cell, where and when it interacts with its binding partners, how it is distributed, and how this is regulated, all at the level of single molecules in living cells. Researchers: Annya Smyth, Kirsty Martin, Colin Rickman, Rory Duncan, Charlotte Hamilton.
Imaging large cohorts of single molecules in 3-Dimensions (Royal Society, to 2013)
Current single molecule imaging approaches (2011), although extremely impressive, are limited in the main to 2-D, often at the cell membrane. This project aims to use the optical tools developed by Professor Alan Greenaway (HWU, Physics) to permit the axial ranging of molecules in 3-D. In short, we expect to be able to localise the positions of single molecules with an accuracy of <20 nm in the XY plane, and <50 nm in Z. This is an enormous improvement on the current state-of-the-art, will be compatible with any commercial fluorescence microscope, and is relatively simple and inexpensive. Researchers: Rory Duncan, Alan Greenaway
Secretory Vesicles (STFC, to 2013)
This project utilises novel imaging approaches being developed by Professor Alan Greenaway (Physics, HWU), to increase the rate of acquisition of 3-D stacks using a widefield microscope. This is important, because our present acquisition rate, of approximately 1 entire cell stack per second, is too slow to permit accurate tracking of fast-moving organelles. By using Alan’s novel optical techniques, we can increase the rate 3, 6 or perhaps even 9-fold - this will permit real-time, 4-dimensional analyses of live-cell dynamics. Researchers: Rory Duncan, Alan Greenaway, PDRA tba
Bacterial protein transport (SULSA, to 2015)
We are applying advanced molecular imaging approaches (FLIM, FCS, TIRFM, PALM) to probe protein interactions in single bacteria. This SULSA-funded project is in collaboration with Professor Frank Sargent, University of Dundee. How bacteria transport proteins between intracellular compartments or out of the cell remains speculative. This project will analyse specific components of the Tat pathway in E. coli to address this. Researchers: Frank Sargent, Kat McGowan, Colin Rickman, Rory Duncan


Our projects all aim to either improve the acquisition of molecular image data from cells, or use those data to answer fundamental questions in cell biology by quantifying the dynamic behaviours of single molecules inside cells.
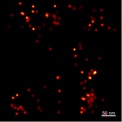
Single molecule analysis of the molecular machinery of regulated secretion (Royal Society, to 2011)
This project examines the spatial distribution of SNARE molecules in neuroendocrine cells and their organisation with relation to secretory vesicles. By utilising the PALM super-resolution technique, we can examine at the individual molecule level large populations of plasma membrane resident SNARE proteins. Through statistical analysis of this data we aim to generate a molecular map of the secretory machinery immediately preceding regulated exocytosis. Researcher: Colin Rickman
LSI Projects
Images and data copyright LSI Laboratory
